Our manuscript describing the dynamics of induced motor neuron programming has been published in Cell Stem Cell. The manuscript, “A multi-step trans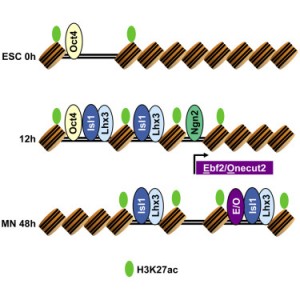 criptional and chromatin state cascade underlies motor neuron programming from embryonic stem cells“, results from a close collaboration between our lab, Esteban Mazzoni’s lab at NYU, and Uwe Ohler’s lab at the Max Delbrück Center for Molecular Medicine in Berlin.
criptional and chromatin state cascade underlies motor neuron programming from embryonic stem cells“, results from a close collaboration between our lab, Esteban Mazzoni’s lab at NYU, and Uwe Ohler’s lab at the Max Delbrück Center for Molecular Medicine in Berlin.
In this study, we studied the process by which mouse embryonic stem cells can be directly (and efficiently) converted to spinal motor neurons via the forced expression of three transcription factors: Ngn2, Isl1, and Lhx3. This direct programming system is uniquely efficient; over 90% of cells are converted to motor neurons within 48 hours. Our system can therefore serve as model of direct cellular programming, allowing us to ask how expression of a few key transcription factors enables conversion of cellular identity. We characterized the dynamics of transcriptional and chromatin changes associated with motor neuron programming using several genome-wide assays including single-cell RNA-seq, bulk RNA-seq, ATAC-seq, and ChIP-seq targeting various transcription factors and chromatin modifications.
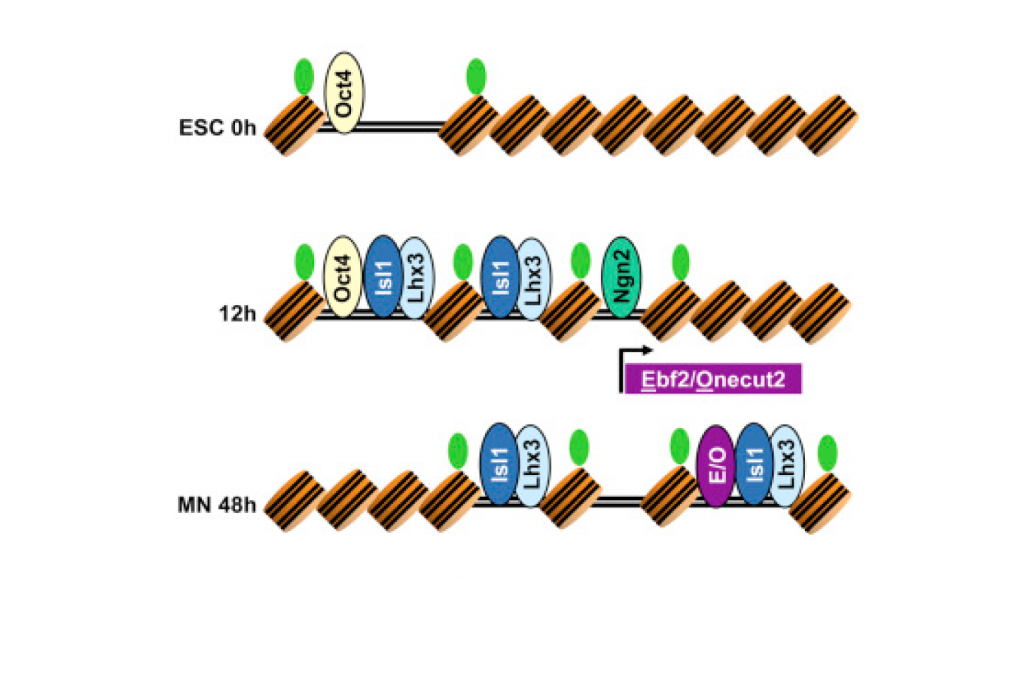
Our results show that programming of ES cells into motor neurons is the result of two independent transcriptional processes that eventually converge. As we previously characterized, two of the transcription factors — Isl1 and Lhx3 — bind together to the genome. Early in the programming process, this pair of factors binds to both regions of the genome that serve as enhancers in ES cells as well as regions that are devoid of regulatory activity. The third transcription factor, Ngn2, acts independently, making additional changes to gene expression, but mainly binds to regions that are already accessible/active in ES cells. Later in the transformation process, Isl1 and Lhx3 shift their binding away from ES enhancers and towards a new set of previously inaccessible sites. This binding shift brings regulatory proteins to key motor neuron identity genes, and appears to be required for direct programming to successfully achieve cellular conversion.
Besides characterizing these interesting dynamics in transcription factor binding, we were able to predict that the shift in Isl1 and Lhx3 binding late in the programming process is dependent on additional transcription factors that are induced by Ngn2. Specifically, factors from the Ebf and Onecut protein families appear to assist Isl1 and Lhx3 in binding to these late control regions. Thus, motor neuron programming is the product of two initially independent transcriptional modules that converge with a feedforward transcriptional logic.
The published manuscript has three joint first authors: Silvia Velasco in the Mazzoni lab, Mahmoud Ibrahim in the Ohler lab, and Akshay Kakumanu in our lab.
Press relating to this work:
Update: This work has been featured in a preview by Cave & Socanathan in the journal Neuron.